 NA-MIC Project Weeks
NA-MIC Project Weeks
Back to Projects List
Review of segmentation results quality across various multi-organ segmentation models
Key Investigators
- Andrey Fedorov (BWH, USA)
- Ron Kikinis (BWH, USA)
- David Clunie (Pixelmed Publishing, USA)
- Steve Pieper (Isomics Inc, USA)
- Andres Diaz-Pinto (NVIDIA, UK)
- Tamaz Amiranashvili (University of Zurich, Switzerland)
- Murong Xu (University of Zurich, Switzerland)
- Klaus Maier-Hein (DKFZ, Germany)
- Bjoern Menze (University of Zurich, Switzerland)
Project Description
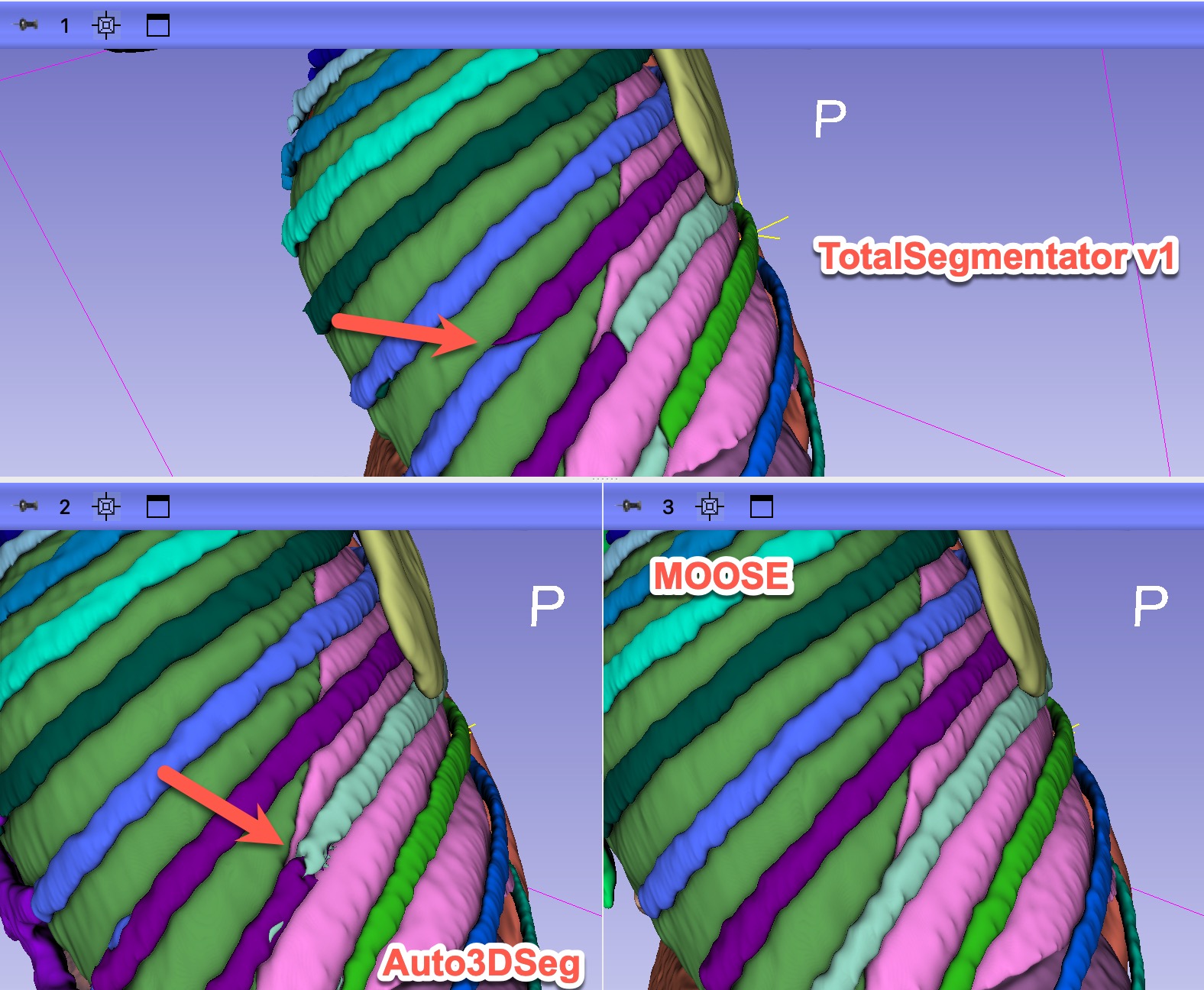
When initially released, TotalSegmentator was perceived to produce superior results, in comparison to the state-of-the-art at the time, anyway.
Over time, some of the deficiencies in the segmentations produced by TotalSegmentator have been identified. Further, new multi-organ segmentation models have been introduced.
Objective
- Review segmentation results for a sample of images from IDC NLST collection, documenting the problems, across the publicly available multi-organ segmentation models.
Approach and Plan
- Identify set of cases to review.
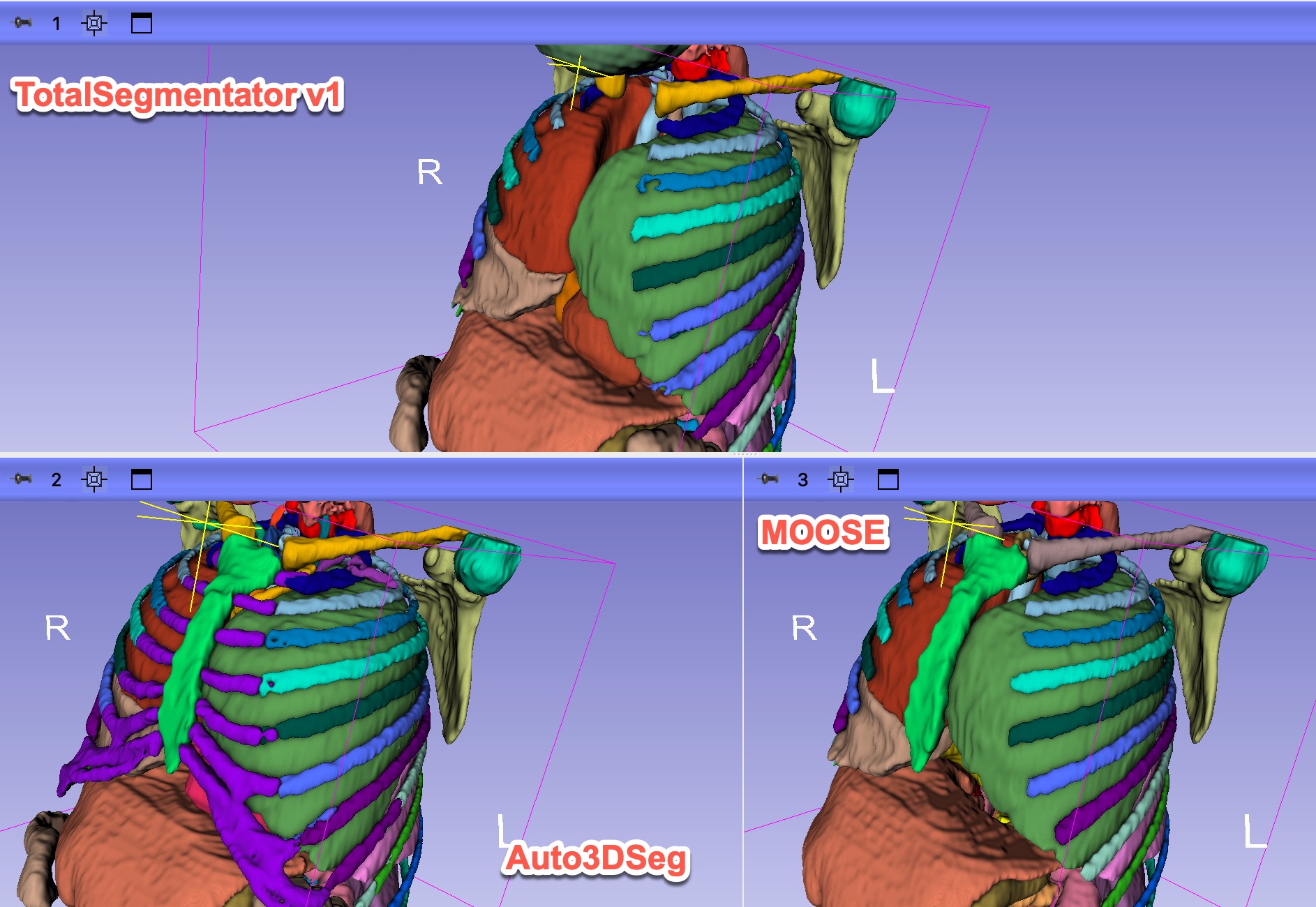
- Collect results from various methods (currently have for MOOSE and TotalSegmentator).
- Review cases with Ron and David (using SegmentationVerificationModule
- Identify more cases with failures, prepare interface/instructions how to help in identifying those.
- Summarize the results of the review in a publicly available document.
Progress and Next Steps
- Instructions for downloading initial sample of images:
pip install --upgrade idc-index
test_series = \
['1.2.840.113654.2.55.195946682403058845904768502826466194287', \
'1.2.840.113654.2.55.221581533879834196356530174246594024639', \
'1.2.840.113654.2.55.71263399928421039572326605504649736531', \
'1.2.840.113654.2.55.79318439085250760439172236218713769408', \
'1.2.840.113654.2.55.191661316001774647835097522264785668378', \
'1.2.840.113654.2.55.304075689731327662774315497031574106725', \
'1.2.840.113654.2.55.283399418711252976131557177419186072875', \
'1.2.840.113654.2.55.21461438679308812574178613217680405233', \
'1.2.840.113654.2.55.97114726565566537928831413367474015470', \
'1.2.840.113654.2.55.122344168497038128022524906545138736420', \
'1.2.840.113654.2.55.229650531101716203536241646069123704792', \
'1.2.840.113654.2.55.257926562693607663865369179341285235858', \
'1.3.6.1.4.1.14519.5.2.1.7009.9004.135383252566920035150987356231', \
'1.3.6.1.4.1.14519.5.2.1.7009.9004.315696884435641630605419115484', \
'1.3.6.1.4.1.14519.5.2.1.7009.9004.230644512623268816899910856967', \
'1.3.6.1.4.1.14519.5.2.1.7009.9004.330739122093904668699523188451', \
'1.3.6.1.4.1.14519.5.2.1.7009.9004.690272753571338193252806012518', \
'1.3.6.1.4.1.14519.5.2.1.7009.9004.310718458447911706151879406927']
from idc_index import IDCClient
c= IDCClient()
c.download_from_selection(downloadDir=".",seriesInstanceUID=test_series)
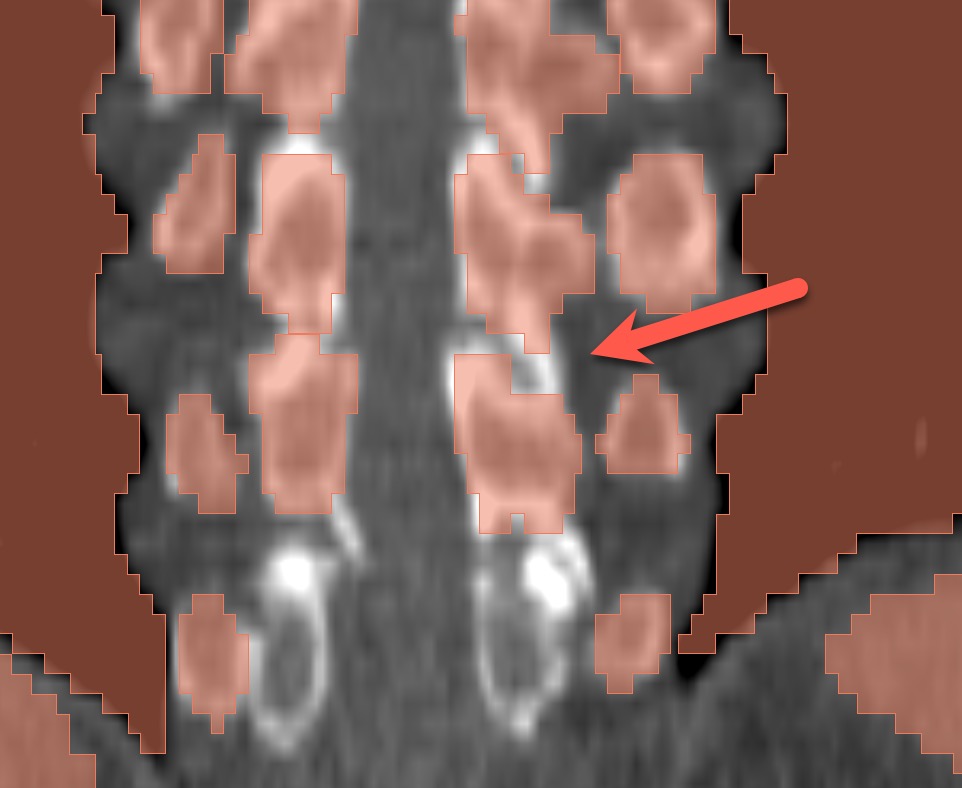
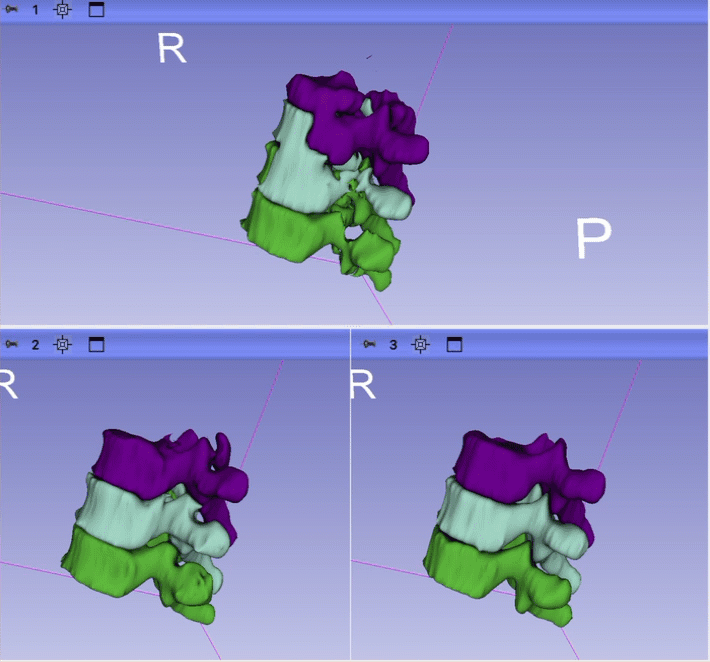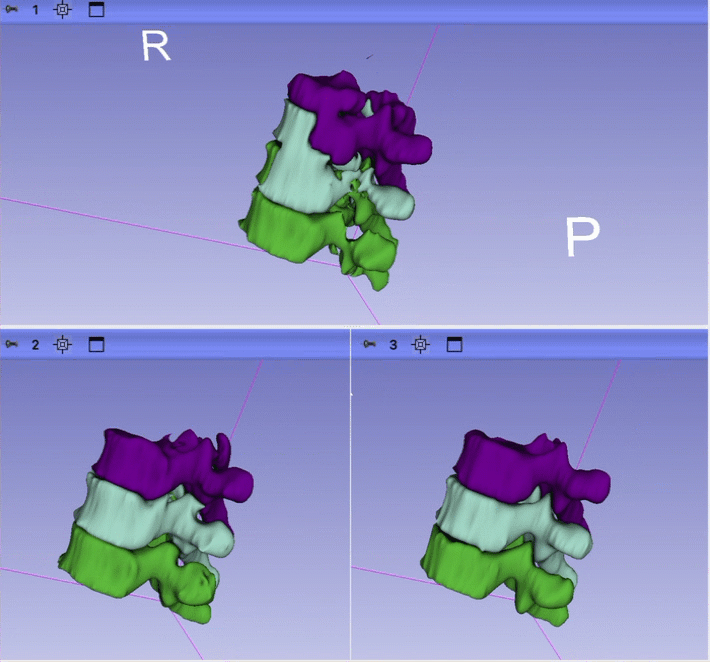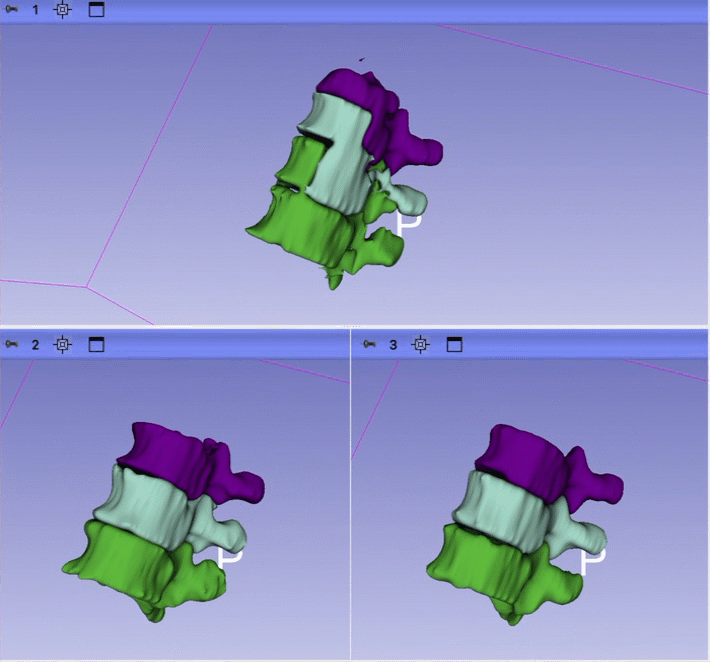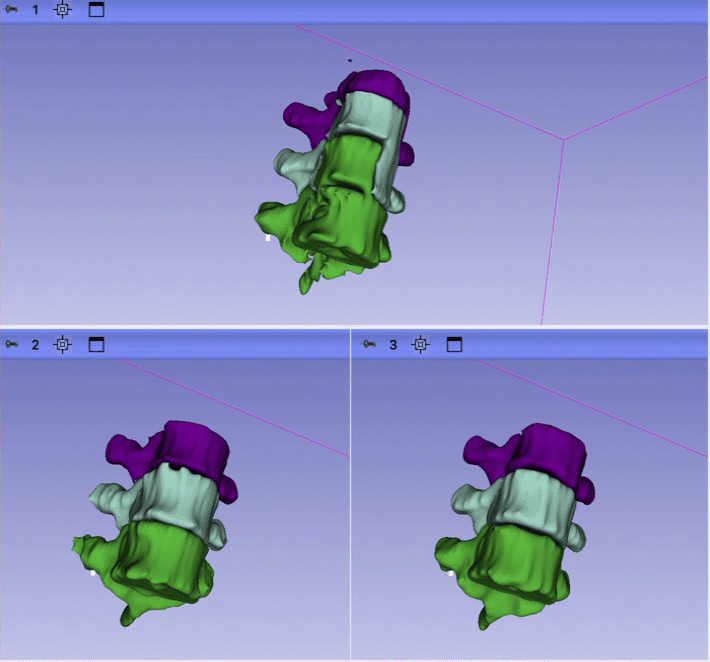
- 1 session with Ron looking at the initial result for just one case. Issues identified: 1) unrealistic anatomy and large gap between ribs and and erector spinae; 2) incorrect segmentation of some vertebrae; 3) large gaps between the structure; 4) segmentation is too coarse.
- Collected results from Auto3DSeg and OMAS, in addition to TotalSegmentator v1 and MOOSE for the test sample.
- Finished SNOMED mapping for OMAS.
- Harmonized mapping from model-specific labels to SNOMED-CT codes and consistent colors for Auto3DSeg and MOOSE (see CSV files here) - Google Sheet with current mapping. Didn’t finish this for OMAS! Notebook for this harmonization here.
- Explored Slicer capabilities for joint visualization of the segmentation results, identified relevant features thanks to Steve!
- Received Multitalent model segmentation results from Klaus Maier-Hein.
- Co-authored with Perplexity a Google Apps script for coloring Google Sheets cells based on the RGB string - helpful for quickly evaluating color selection.
- Brainstormed ideas for how to approach and scale up this comparison with Bjoern.
function colorCellsFromRGB() {
var sheet = SpreadsheetApp.getActiveSpreadsheet().getActiveSheet();
var dataRange = sheet.getDataRange();
var values = dataRange.getValues();
for (var i = 0; i < values.length; i++) {
for (var j = 0; j < values[i].length; j++) {
var cell = values[i][j];
if (typeof cell === 'string' && cell.match(/^\[\d{1,3}, \d{1,3}, \d{1,3}\]$/)) {
var rgb = JSON.parse(cell);
var color = rgbToHex(rgb[0], rgb[1], rgb[2]);
sheet.getRange(i + 1, j + 1).setBackground(color);
}
}
}
}
function rgbToHex(r, g, b) {
return "#" + ((1 << 24) + (r << 16) + (g << 8) + b).toString(16).slice(1);
}
Feedback/features:
- identified some issues in SegmentationReview module - Andrey will try to submit a PR
- identified issues related to visualization of segmentations in Slicer - issue submitted https://github.com/Slicer/Slicer/issues/8190
- would be really helpful to be able to show what structure cursor points to in 3d View (Ron seconds this!)
- review of multiple segmentations is currently difficult - perhaps opportunity for improving SegmentationsReview module
Illustrations
No response
Background and References
- Krishnaswamy, D., Thiriveedhi, V. K., Ciausu, C., Clunie, D., Pieper, S., Kikinis, R. & Fedorov, A. Rule-based outlier detection of AI-generated anatomy segmentations. arXiv [eess.IV] (2024). at http://arxiv.org/abs/2406.14486
- HuggingFace exploration dashboard: https://huggingface.co/spaces/ImagingDataCommons/CloudSegmentatorResults