 NA-MIC Project Weeks
NA-MIC Project Weeks
Back to Projects List
Evaluation of imi-bigpicture/wsidicomizer as a tool for conversion into DICOM whole slide imaging format
Key Investigators
- Andrey Fedorov (BWH, USA)
- Daniela Schacherer (Fraunhofer MEVIS, Germany)
- David Clunie (Pixelmed Publishing, USA)
- Max Fischer (DKFZ, Germany)
Project Description
DICOM standard is gaining acceptance in digital pathology imaging. Conversion of slide images into DICOM format can make the data more FAIR, improve quality and comprehensiveness of the associated metadata, and improve their interoperability with the commercial and open source tools implementing the standard.
DICOM format is used for slide microscopy images available in NCI Imaging Data Commons (IDC). Images submitted to IDC in the vendor-specific formats must be converted into DICOM representation, which is currently done using the PixelMed Toolkit based scripts available in https://github.com/ImagingDataCommons/idc-wsi-conversion.
Our goal is to migrate the DICOM WSI conversion to use community-supported open source tools. Based on our current assessment and experience, imi-bigpicture/wsidicomizer is the most promising tools available for this task. In this project we will work on evaluating this tool.
Objective
- Assemble inventory of the publicly available test samples representative of the variety of data encountered by IDC and perhaps outside of IDC.
- Document requirements for the conversion tool based on the needs of IDC.
- Complete evaluation of
wsidicomizerand document the results (in terms of the features and performance of the conversion process). - Document results and identified gaps to help with the next steps.
Approach and Plan
- Select representative source images in the original format and the results of conversion to DICOM available in IDC (as converted using PixelMed Toolkit), as a reference. Assemble information about the characteristics of those samples in a document (vendor, compression, …). Include the accompanying tabulated metadata that is needed for converting each particulare sample.
- Requirements: initialization of metadata, standard compliance of the result, transfer of ICC profile, acceptable performance …. (intentionally, DICOM-TIFF dual personality at this point is not a requirement)
- Create a publicly available script/notebook that performs conversion.
- Evaluate the results and summarize in a publicly available document.
- Document any identified problems by opening issues in the
wsidicomizerrepo.
Progress and Next Steps
- Set up conversion code in python (simple), confirmed conversion approach is consistent between what we use in IDC and what Max is using in Kaapana (
wsidicomizerPython function - not command line tool). - Prepared queries for selecting test images from IDC. Mapping to the source file in vendor format is stored in a private tag
(0009,1001)(source non-DICOM files are in private buckets in IDC). - Identified problems in selecting samples based on
TransferSyntaxUID- did not realize initially it can vary across instances within the same series! - Identified numerous very strange images in IDC - will need to investigate this further.
- Started testing
wsidicomizer, tested with JPEG and uncompressed samples. - Identified and reported converter issues, several of which have already been resolved (kudos to Erik Gabrielsson,
wsidicomizermaintainer!):- https://github.com/imi-bigpicture/wsidicomizer/issues/117
- https://github.com/imi-bigpicture/wsidicomizer/issues/118
- https://github.com/imi-bigpicture/wsidicomizer/issues/123
- Discussed various issues related to conversion and shared experience; reached agreement
wsidicomizeris the best choice given combined experience, and very good support from Erik. - Identified issues in
dicom3toolsbuilding it in Colab VM - fixed by David Clunie (notebook).
Query for selecting samples from IDC based on TransferSyntaxUID applied to the base layer of the image pyramid:
WITH
RankedRows AS (
SELECT
SeriesInstanceUID,
StudyInstanceUID,
TotalPixelMatrixColumns*TotalPixelMatrixRows AS totalPixels,
TransferSyntaxUID,
ROW_NUMBER() OVER (PARTITION BY SeriesInstanceUID ORDER BY TotalPixelMatrixColumns*TotalPixelMatrixRows DESC) AS rn
FROM
`bigquery-public-data.idc_current.dicom_all`
WHERE
Modality = "SM" and collection_id not like "%htan%")
SELECT
TransferSyntaxUID,
StudyInstanceUID,
SeriesInstanceUID,
totalPixels,
concat("https://viewer.imaging.datacommons.cancer.gov/slim/studies/",StudyInstanceUID,"/series/",SeriesInstanceUID)
FROM
RankedRows
WHERE
rn = 1
# Explicit VR Little Endian
AND TransferSyntaxUID = "1.2.840.10008.1.2.1"
ORDER BY
totalPixels ASC
Illustrations
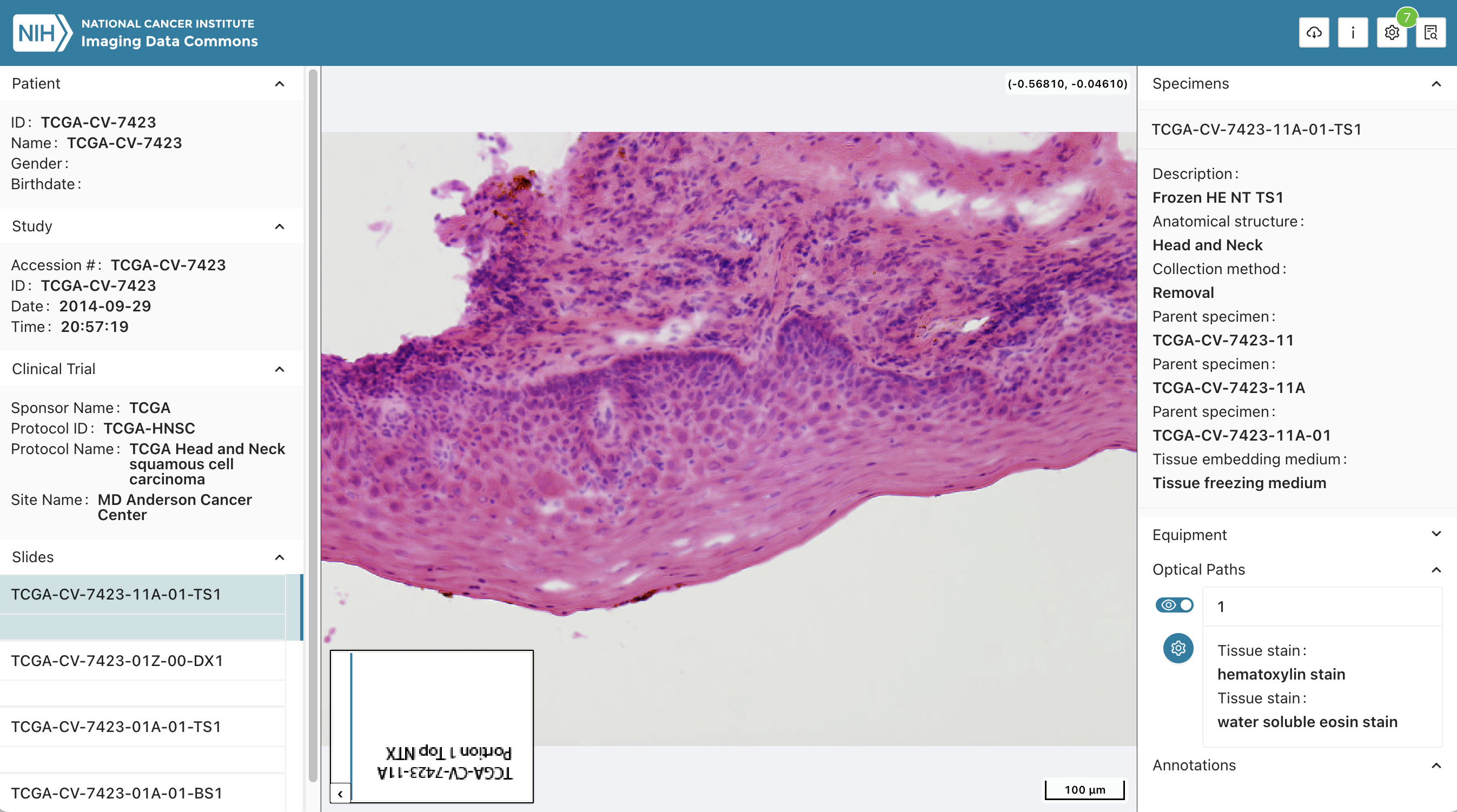
Background and References
Background reading:
- Herrmann, M. D., Clunie, D. A., Fedorov, A., Doyle, S. W., Pieper, S., Klepeis, V., Le, L. P., Mutter, G. L., Milstone, D. S., Schultz, T. J., Kikinis, R., Kotecha, G. K., Hwang, D. H., Andriole, K. P., John Lafrate, A., Brink, J. A., Boland, G. W., Dreyer, K. J., Michalski, M., Golden, J. A., Louis, D. N. & Lennerz, J. K. Implementing the DICOM standard for digital pathology. J. Pathol. Inform. 9, 37 (2018). http://dx.doi.org/10.4103/jpi.jpi_42_18
- Clunie, D. A. DICOM format and protocol standardization-A core requirement for digital pathology success. Toxicol. Pathol. 49, 738–749 (2021). http://dx.doi.org/10.1177/0192623320965893
Other related materials:
- IDC Portal: https://portal.imaging.datacommons.cancer.gov/
- conversion_mirax_dicom (currently, private repo)
- Related earlier project from PW40: https://projectweek.na-mic.org/PW40_2024_GranCanaria/Projects/WsiDicomImprovementFromViewerToAnalysis/
- Code from Fabian Hörst experiments: https://github.com/TIO-IKIM/PathoPatcher/blob/main/pathopatch/patch_extraction/patch_extraction.py
- Test samples used by wsidicomizer: https://github.com/imi-bigpicture/wsidicomizer/blob/main/tests/download_test_images.py#L23-L60
- Google WSI transformation pipeline