NA-MIC Project Weeks
NA-MIC Project Weeks
Back to Projects List
Deploying OvSeg in Slicer
Key Investigators
- Paolo Zaffino (Magna Graecia University of Catanzaro, Italy)
- Thomas Buddenkotte (University Medical Center HamburgEppendorf, Germany)
Project Description
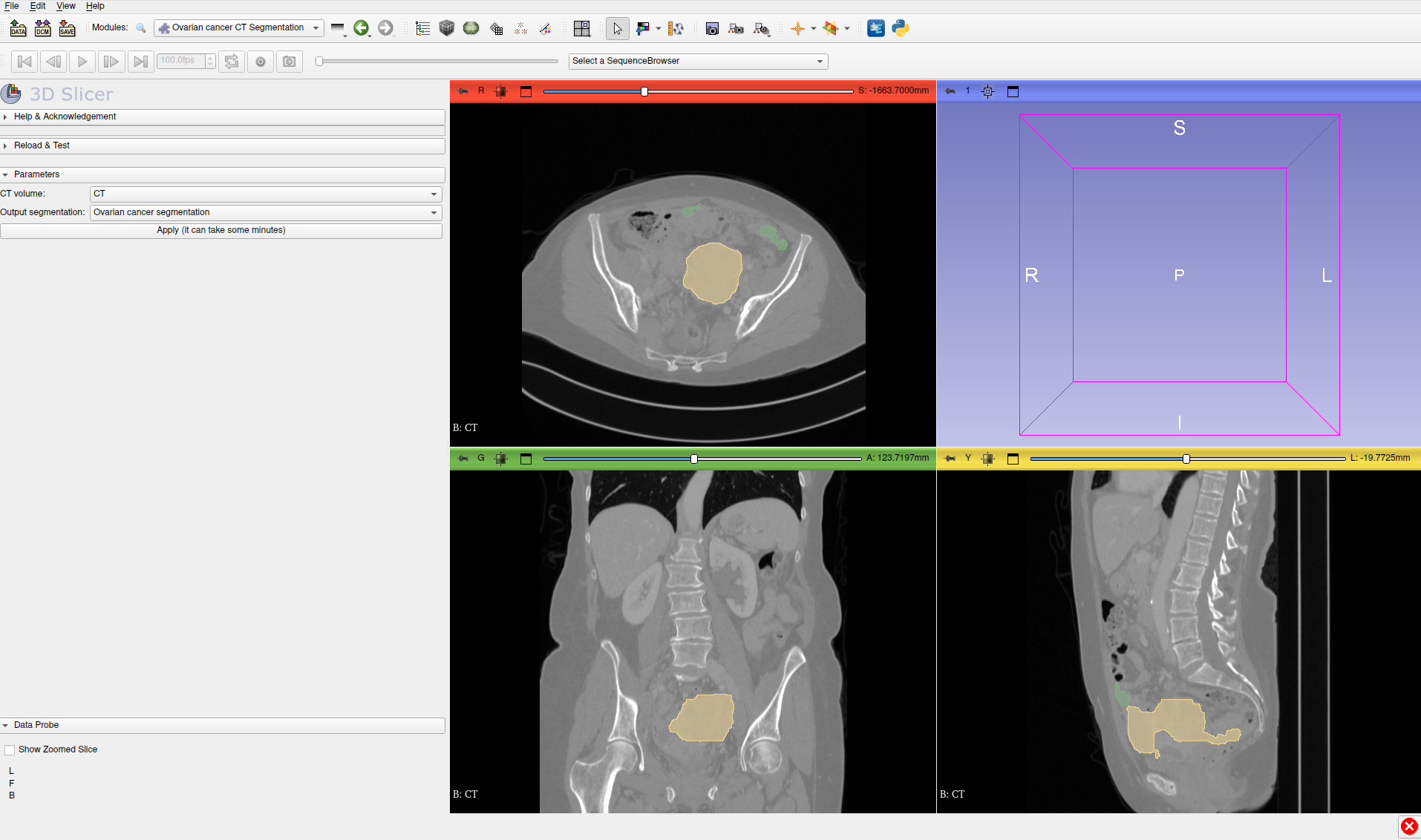
OvSeg is a deep learning-based library for the segmentation of high-grade serous ovarian cancer on CT images. Right now, to obtain the segmentations the user has to write some lines of Python code, making the tool not directly usable by non-technical people. It would be great to expose this algorithm in Slicer to be used in a codeless manner.
Objective
- Expose OvSeg in 3D Slicer in order to provide a scalar volume as input and obtain the segmentations as a segmentation node
Approach and Plan
- Create a Slicer extension
- Let the extension to install OvSeg via pip
- Let the extension pull the CT volume, run the inference, and push back the segmentations
Progress and Next Steps
- Ovseg can be successfully used in Slicer
-
The code can be downloaded from this GitHub repository
- We would like to include this extension in the Slicer Extension repository
- We need a logo
- We could decide to expose the extended ovseg version which provides more segmentations classes
Illustrations